| tfrmt_sigdig {tfrmt} | R Documentation |
Create tfrmt object from significant digits spec
Description
This function creates a tfrmt based on significant digits specifications for
group/label values. The input data spec provided to sigdig_df will contain
group/label value specifications. tfrmt_sigdig assumes that these columns
are group columns unless otherwise specified. The user may optionally choose
to pass the names of the group and/or label columns as arguments to the
function.
Usage
tfrmt_sigdig(
sigdig_df,
group = vars(),
label = quo(),
param_defaults = param_set(),
missing = NULL,
tfrmt_obj = NULL,
...
)
Arguments
sigdig_df |
data frame containing significant digits formatting spec.
Has 1 record per group/label value, and columns for relevant group and/or
label variables, as well as a numeric column |
group |
what are the grouping vars of the input dataset |
label |
what is the label column of the input dataset |
param_defaults |
Option to override or add to default parameters. |
missing |
missing option to be included in all |
tfrmt_obj |
an optional tfrmt object to layer |
... |
These dots are for future extensions and must be empty. |
Details
Formats covered
Currently covers specifications for frmt and
frmt_combine. frmt_when not supported and must be supplied in additional
tfrmt that is layered on.
Group/label variables
If the group/label variables are not provided to the arguments, the body_plan will be constructed from the input data with the following behaviour:
If no group or label are supplied, it will be assumed that all columns in the input data are group columns.
If a label variable is provided, but nothing is specified for group, any leftover columns (i.e. not matching
sigdigor the supplied label variable name) in the input data will be assumed to be group columns.If any group variable is provided, any leftover columns (i.e. not matching
sigdigor the supplied group/label variable) will be disregarded.
Value
tfrmt object with a body_plan constructed based on the
significant digits data spec and param-level significant digits defaults.
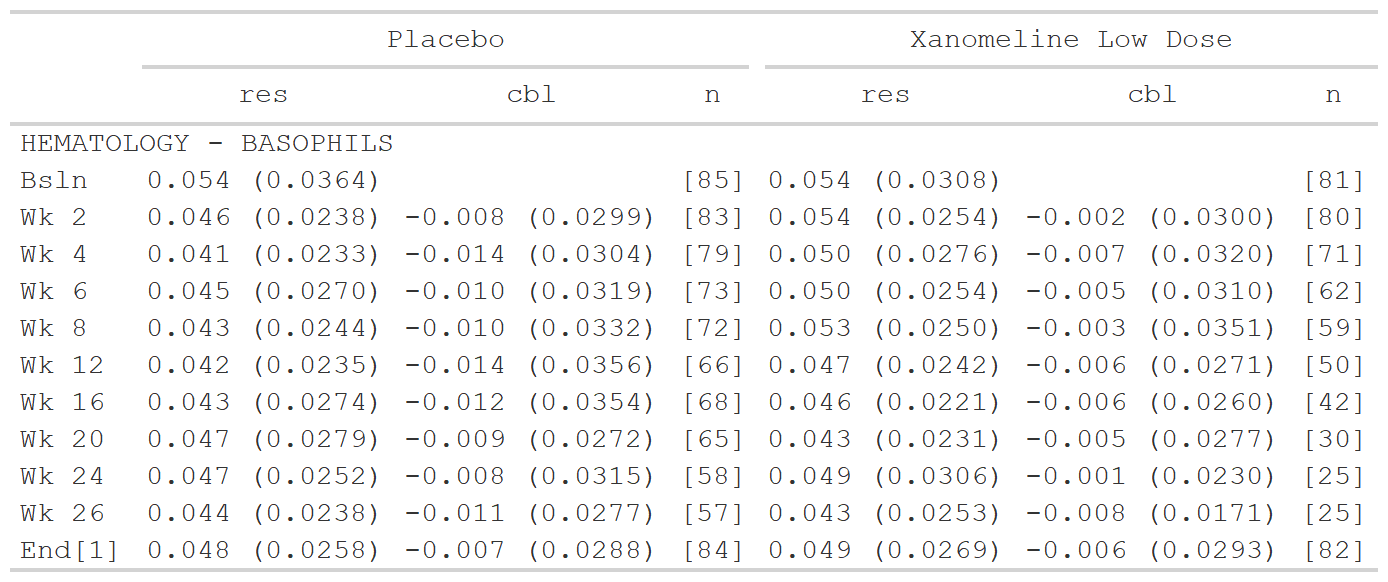
Examples
sig_input <- tibble::tribble(
~group1, ~group2, ~sigdig,
"CHEMISTRY", ".default", 3,
"CHEMISTRY", "ALBUMIN", 1,
"CHEMISTRY", "CALCIUM", 1,
".default", ".default", 2
)
# Subset data for the example
data <- dplyr::filter(data_labs, group2 == "BASOPHILS", col1 %in% c("Placebo", "Xanomeline Low Dose"))
tfrmt_sigdig(sigdig_df = sig_input,
group = vars(group1, group2),
label = rowlbl,
param_defaults = param_set("[{n}]" = NA)) %>%
tfrmt(column = vars(col1, col2),
param = param,
value = value,
sorting_cols = vars(ord1, ord2, ord3),
col_plan = col_plan(-starts_with("ord"))) %>%
print_to_gt(.data = data)